Features
Model-Based (Model Fitting) Analysis Method
The Unique Model-Based Kinetics for Comprehensive Analysis of Chemical Reactions
Approximately 95% of all chemical reactions are multi-step reactions. This requires a multi-step analytical engine as offered by the Kinetics Neo software.
Model-based (model fitting) method of kinetic analysis is the analysis method for complex chemical process consisting of individual reaction steps, where each step can be individually connected to another reaction steps (consecutive, competitive, independent) in order to build kinetic model of complex process. Model-based method describe reaction rate of multi-step chemical reactions by the system of kinetic equations where each reaction step has own kinetic equation and own kinetic triplet containing activation energy, pre-exponential factor A and reaction type.
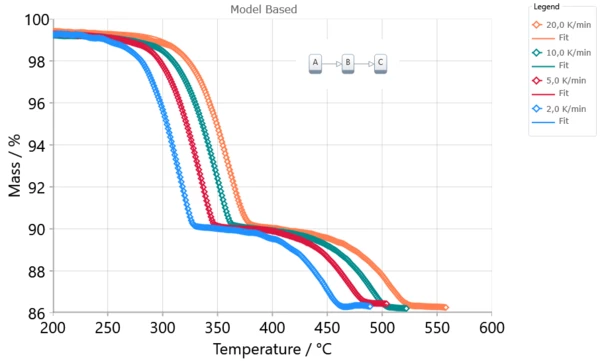
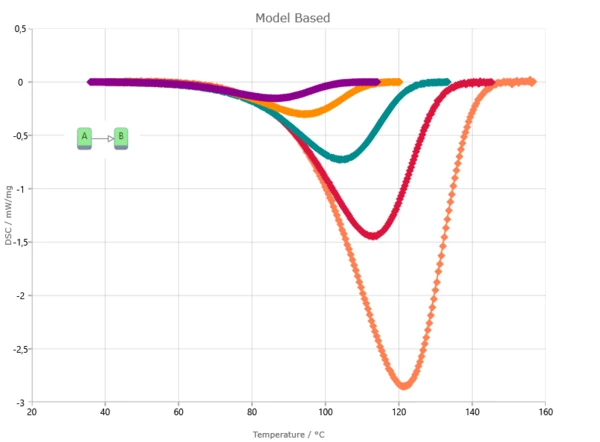
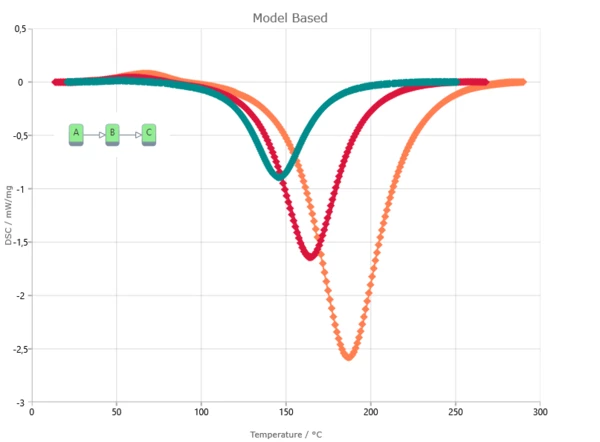
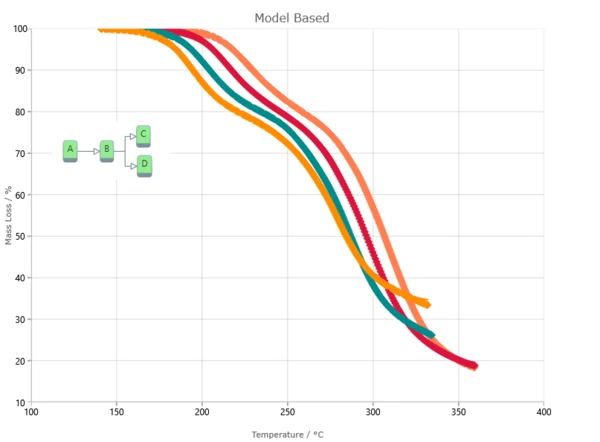
Result of model-based method is the kinetic model with kinetic parameters for each reaction step. Kinetic parameters are found from the best fit of kinetic model for experimental data. Model-based method can show reaction rate for each step and concentration of each reactant.
The extraordinary model-based analysis was developed by NETZSCH. It uses powerful cutting-edge mathematical calculations to create the best kinetic model; the different kinetic models can then also be compared statistically. Therefore, this approach has none of the disadvantages which can be observed when using model-free methods.
The Model-Based Kinetic Analysis Is Based on Three Assumptions:
1. The reaction consists of several elementary reaction steps, and the reaction rate of each step can be described by a kinetic equation of its own for the given step, depending on the concentration of the initial reactant ej, the concentration of product pj, the pre-exponential factor Aj and the activation energy Ej, specific only for this step with number j, as follows:
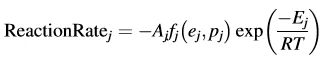
Each step has its own reaction type described by the function fj (ej;pj).
Some examples of these functions include a second-order reaction which has f = e2, a Prout–Thompkins reaction with acceleration which has f = empn and a reaction with a one-dimensional diffusion which has f = 0.5/p. The number of kinetic equations is equal to the number of reaction steps; the concentration for each reactant increases for the reaction steps where this reactant is a product, and decreases for the reaction steps where this reactant is a starting substance.
2. All kinetic parameters including the activation energy, pre-exponential factor, order of reaction, and reaction type are assumed to be constant during the reaction progress for every individual reaction step.
3. The total thermoanalytical signal is the sum of the signals of the individual reaction steps. The signal of each step is calculated as the reaction rate multiplied by the total effect of the given step; e.g., total enthalpy change or total mass loss.
Unique Flexible Model Designer in Kinetics Neo
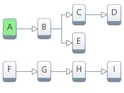
Model-based kinetic analysis offers the possibility of visual design for kinetic models with an unlimited number of steps connecting in any combinations.
The models can be flexibly designed by adding new reactions as independent, consecutive or competitive steps to any place in the model.
A simulated reaction step can be visually moved to the corresponding step on the experimental curve. Then the parameters of this step can be optimized.
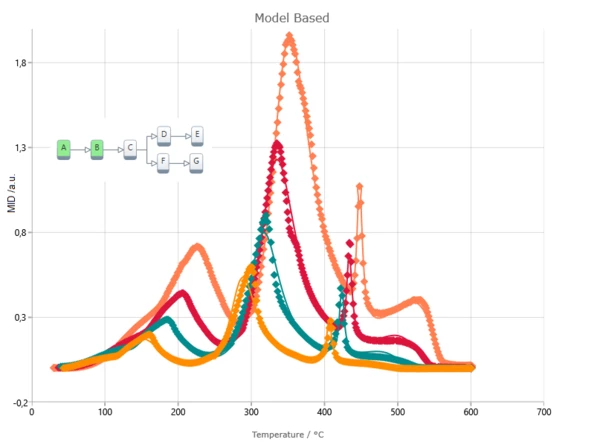
Model-Based Method Results
The Kinetics Neo engine uses non-linear regression methods and allows for the optimization of parameters for individual steps or for the complete model. The fit results present the agreement between the experimental and simulated curves for:
- Signal
- Conversion
- Conversion rate
- Concentration of all reactants
- Reaction rates for all steps